- General Questions
- Usability Questions
General Questions
What is boostDM?
boostDM is a computational method to conduct in silico saturation mutagenesis of cancer genes. Using mutational features, the method scores and classifies all possible single base substitutions in cancer genes by their potential to be involved in tumorigenesis.
How does it work?
The method is based on the analysis of observed mutations in sequenced tumors and their site-by-site annotation with relevant features. The compendium of cancer genes and the mutational features for each cancer gene across malignancies have been derived from the systematic analysis of tens of thousands of tumor samples (www.intogen.org). Other relevant features have been collected from public databases.
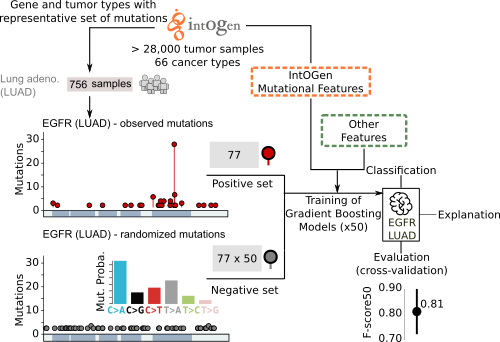
What is the main novelty?
The main novelty of boostDM is that it builds one independent model for every cancer gene-tumor type combination. This has far-reaching implications in the way the model output is interpreted and used for downstream inquiry. Additionally, it does not require any pre-annotated labelling of driver or passenger mutations. Instead, boostDM draws both driver and passenger training mutations from the same genes harbouring the cancer driver signals. We also aimed to make classification of each possible mutation in a cancer gene readily interpretable. For that purpose, the features used to train the models of each cancer gene are mutational patterns that are closely connected to the signals of positive selection discerned (IntOGen) across cohorts of tumors.
How does boostDM training work?
boostDM resorts to supervised learning using a training dataset made of "driver" and "passenger" mutations from a cancer gene. By means of dN/dS analysis (dNdScv), we can establish a proportion of mutations observed that cannot be explained by neutral evolution (excess). To train boostDM, we include in the "driver" training set all the mutations observed in cancer driver genes (IntOGen) with high enough excess (≥ 85%). Since the excess can be estimated in a consequence-type specific manner, we can include only those mutations with consequence types having high excess. On the other hand, the "passenger" training set are mutations randomly drawn with flat probabilities based on the tri-nucleotide context rates found in the tumor type cohort.
Where do you get the data from?
boostDM models feed on IntOGen (currently from release 2020-02-01).
What features do the models use?
Each mutation provided for training is annotated with a set of mutational features, which the classification task exploits to discriminate between observed drivers and passengers in tumours. Some mutational features have been derived from the systematic analysis of tens of thousands of tumor samples from IntOGen (e.g., linear clusters, 3D clusters, enriched functional domains). Other relevant features, such as nucleotide conservation or post-translational modifications are collected from public databases of biological sequences. Here we introduce a brief description of the features the user will find in this site. For a more detailed account, please refer to the manuscript.
- SYNONYMOUS, MISSENSE, NONSENSE: whether the mutation induces any of these consequence types at the protein level (ENSEMBL Variant Effect Predictor).
- SPLICE: whether the mutation has the potential to disrupt RNA splicing in the canonical transcript (ENSEMBL Variant Effect Predictor).
- ACETYLATION, METHYLATION, PHOS (phosphorylation), REG (regulatory site), UB (ubiquitination): whether the mutation maps to a protein site known to undergo post-translational modification (PhosphoSitePlus).
- 3CT, 3C: whether the mutation maps to a mutational cluster in the 3D protein conformation (HotMaps3D) in the tumor type and in other tumor types, respectively.
- LCT, LC: whether the mutation maps to a linear cluster (oncodriveCLUSTL) in the tumor type and in other tumor types, respectively.
- LCS: linear cluster score (oncodriveCLUSTL).
- DOM: whether the mutation maps to a PFAM domain that is recurrently mutated in the tumor type (smRegions).
- PHYLOP: nucleotide conservation score asserted through multiple sequence alignments of 99 vertebrate genomes against the human genome (100-way score).
- NMD: whether the (nonsense) mutation is reported in the last exon of the canonical transcript, implying higher chances to undergo inefficient nonsense-mediated mRNA decay.
How do you evaluate the quality of the models?
Each model generated by boostDM is realized as the aggregation of a collection of classifiers trained with partial views of the training data. In the current version 50 classifiers were trained with random partial views of the set of driver and passenger mutations. For each classifier as many passengers as drivers are independently drawn. Then 70% of drivers and passengers are selected for training, whereas the remaining 30% are kept for testing (after removing repeated mutations) consistently with a cross-validation approach. Each classifier attained a test performance that we can measure as an area under the Receiver Operating Characteristic (ROC) curve (auROC). The reported quality of each model is witnessed by the average auROC and by the average number of unique mutations in the test set across the 50 classifiers.
Does every cancer gene have a model?
The long-term goal of boostDM is to come up with a specific model for every cancer gene and every tumor type in which the gene is found to act as a driver according to IntOGen. However, the creation of a reliable model ultimately relies on two hard constraints: i) whether the estimated dN/dS of the gene is high enough to render a training set; ii) whether the number of observed mutations is enough to create a reliable predictor.
What is considered a reliable model?
The cross-validation performance of specific models typically increases with the number of mutations employed to train them. We deem good quality models those trained with ≥ 30 mutations, with average cross-validation F-score50 ≥ 0.8 and such that the observed mutations in that gene and tumor-type are highly representative (see figure below). As more datasets of sequenced tumors become available, we foresee that the number of reliable models will increase.
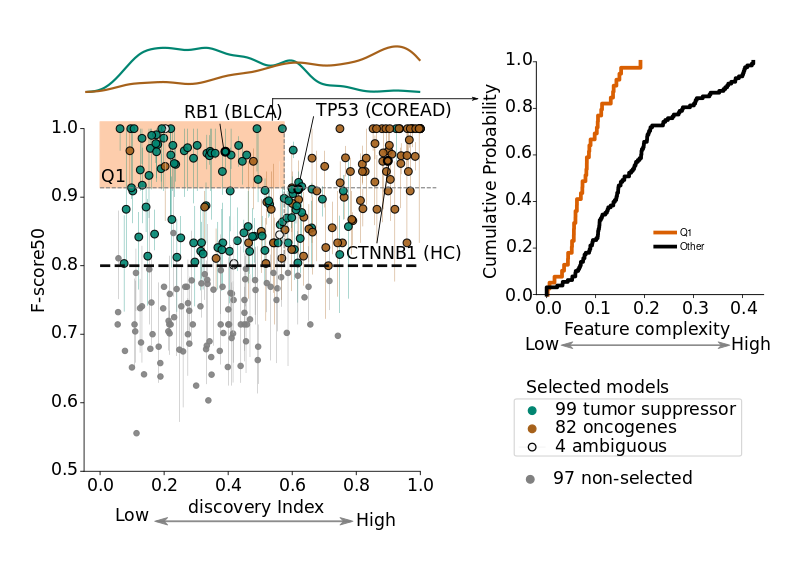
What to do with mutations lacking a specific model?
To classify mutations matching gene-tumor type combinations which do not fulfill the quality requirements described above, we resort to more general models trained with data sets obtained by pooling the mutations of cohorts of related malignancies into meta-cohorts (according to the oncotree ontology). The most suitable non-specific model to classify such mutations is decided on the basis of the first available model (meeting the quality requirements described above) in the oncotree path of decreasing specificity starting in the most specific tumor type that matches the biological context of the mutation (see the Supplementary Note of our manuscript for detailed information about model selection). Notice that even more general models representing shared features of mutations across all tumor suppressors or oncogenes (or all cancer genes) in cohorts or meta-cohorts are also possible. However, the interpretation of these models can be tricky, as these models may be dominated by the mutations in a few highly-mutated genes.
How often are you planning to update the models?
We plan to update the models with every major release of IntOGen. IntOGen is updated regularly with publicly available data from somatic point mutations from patient cohorts. We foresee that as more samples are included in IntOGen, more specific models meeting the minimum quality required will be available.
What gene-tumor type models are featured in the website?
The current version of the web only supports 248 gene-tumor type models: 185 models with highest tumor-type specificity according to our oncotree ontology and 63 additional models where the predictions for a given tumor type required using aggregating the mutations of several tumor types organized in a hierarchical way.
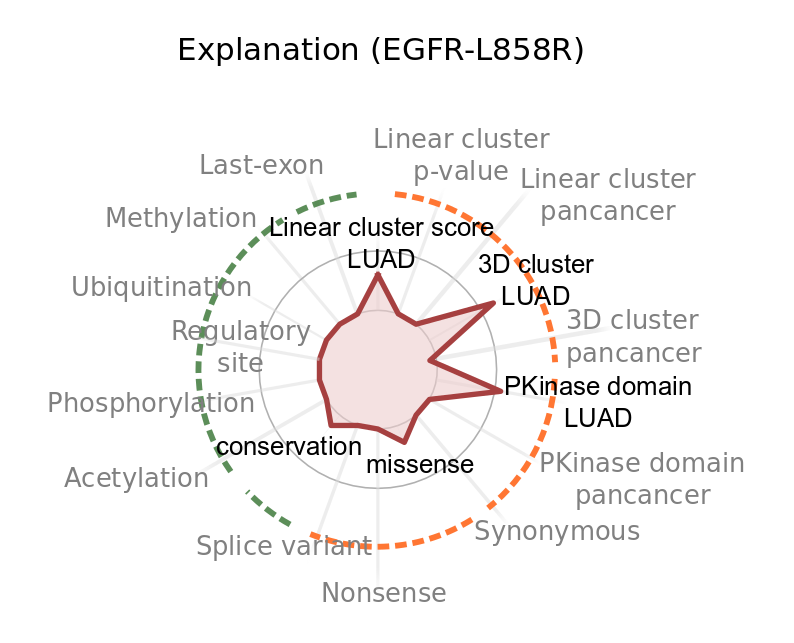
Does boostDM support feature explanations?
The tree-ensemble structure of the gradient boosting classifiers allows the use of Shapley Additive Explanations (SHAP) to infer additive explanation models, i.e. a rule to additively split the forecast produced for each individual mutation in terms of the relative contribution by each feature. More specifically, the SHAP values associated to the features additively break down the logit prediction cast for each input mutation. In particular, SHAP values are to be interpreted in the log-odds scale. Since our models are ensembles of predictors, we report the average SHAP values for the prediction of each individual mutation. Features with higher absolute SHAP values have greater contribution to the classification of a driver mutation. The sign of the SHAP value represents whether the feature value, given the other feature values, implies a higher (positive SHAP) or lower (negative SHAP) probability for the mutation to be a driver.
Have you validated boostDM against experimental datasets?
We conducted an independent evaluation of boostDM against annotated collections of mutations obtained from experimental saturation mutagenesis assays, functional assays and manually curated collections of pathogenic and bening mutations. We reported the results of this analysis in our manuscript.
How do I cite boostDM?
In silico saturation mutagenesis of cancer genes
Ferran Muiños, Francisco Martinez-Jimenez, Oriol Pich, Abel Gonzalez-Perez, Nuria Lopez-Bigas
DOI:10.1038/s41586-021-03771-1
What is the boostDM License?
All data released by this website is available under the Creative Commons Attribution-NonCommercial 4.0 International license. Fair attribution supports future efforts and ensures correct legacy of the data.
What did it take to develop boostDM?
boostDM has come about as a result of a multidisciplinary effort contributed by a team of scientists and engineers in differents areas of expertise: 1) conceptualizing, implementing and testing the learning strategy; 2) conducting validation analyses; 3) processing the data provided by IntOGen; 4) implementing the workflow; 5) implementing the website; 6) preparing the accompanying figures and documentation; 7) maintaining the HPC infrastructure to carry out all the tests and analyses; 8) following-up, putting ideas together and discussing the most suitable analyses and steps forward; 9) coordinating the team efforts.
Who contributed to boostDM?
boostDM is a team effort from the Biomedical Genomics lab (https://bbglab.irbbarcelona.org/) at the Institute for Research in Biomedicine (IRB Barcelona). Ferran Muiños, Francisco Martinez-Jimenez, Oriol Pich, Abel Gonzalez-Perez and Nuria Lopez-Bigas led the main conceptualization, development and validation of the method. We acknowledge technical contributions to the development of boostDM by Iker Reyes-Salazar, Loris Mularoni, Electra Tapanari and Claudia Arnedo-Pac. We also acknowledge the contributions of Miguel L. Grau, David Martínez Millán and Iker Reyes-Salazar in the development of this website.
Usability Questions
What is the current status of the website?
The website is currently a stable alpha version subject to a few minor adjustments. Any feedback or bug reporting is invaluable for us. Please, would you come across any issue, let us know here: bbglab@irbbarcelona.org.
How to read the results?
There are essentially six different types of columns:- Gene information: gene symbol (gene), gene identifier (ENSEMBL_GENE), transcript identifier (ENSEMBL_TRANSCRIPT).
- Mutation descriptors: chromosome (chr), position (pos), alternate allele (alt), amino acid change (aachange).
- Mutational features used to match each mutation to a tumorigenic potential: CLUSTL scores; HotMaps scores; smRegions score (signif_motif); conservation score (PhyloP); last exon (nmd); post-translational modification site annotations (Acetylation, Methylation, Phosphorylation, Regulatory_Site, Ubiquitination); coding consequence type (csqn_type_*).
- Gene and tumor type levels from which the prediction has been drawn (selected_model_gene, selected_model_ttype).
- Predicted tumorigenic potential (boostDM_score, boostDM_class).
- Feature explanation values (SHAP) inferred for each individual prediction (shap_*, following by the name of the mutational feature.
What is the reference genome?
GRCh38/hg38
What transcripts and consequence types are we using?
Only mutations mapping to the canonical transcript according to the ENSEMBL Variant Effect Predictor version 92 (VEP.92) are represented. The consequence of a mutation is consistent with the canonical transcript of the gene. Only mutations with the most damaging consequence being non-synonymous are shown.
What do the tumor type acronyms mean?
You can download the docs describing the tumor type ontology from the downloads page.
What gene-tumor type combinations are included?
First and foremost, only genes classified as drivers by IntOGen in specific tumor types can be valid gene-tumor type combinations. Second, since boostDM learns to evaluate the driver potential of mutations in driver genes from the training examples we feed it on for some gene-tumor type combinations the performance after training may be poor, generally because of the low number of training examples and/or low representability of the mutations observed for that gene and tumor type. We are presenting the results for 248 gene-tumor type combinations which attain a minimum reliability (see What gene-tumor type models are featured in the website? ).
Can I run the code to perform the predictions?
The training and prediction pipeline can be forked from this repo: https://bitbucket.org/bbglab/boostdm/src/rollback/. Please, follow the documentation provided in the repo to satisfy all the data and software requirements.
Can I download the code to perform the predictions?
For the time being it is not possible, but we have the intention to enable this option at some point.
Can I access previous releases of boostDM?
Coming soon
Can I provide feedback?
Yes, definitely, the resource is still undergoing beta testing. Any feedback is invaluable to us. Please, feel free to drop your comments here: bbglab@irbbarcelona.org.
Why does this site use cookies and what for?
We are using Google Analytics cookies to track usage of our site. boostDM is a publicly-funded project and these metrics are important to keep support for this project.