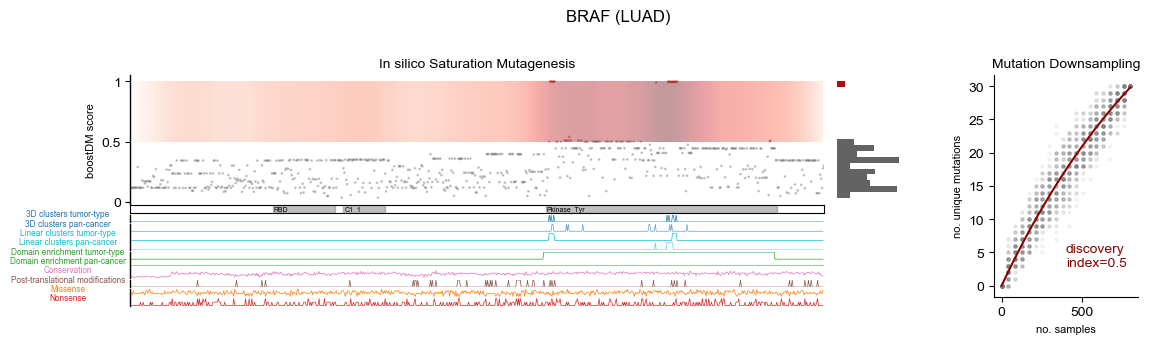
BRAF
LUAD
×
Select a mutation to see further information
| Mutation | AA change | Consequence type | BoostDM score | IntOGen frequency |
|---|---|---|---|---|
| 7:140753336:T | V600E | missense_variant | 1.00 | 10 |
| 7:140781602:A | G469V | missense_variant | 1.00 | 7 |
| 7:140781611:A | G466V | missense_variant | 1.00 | 3 |
| 7:140753393:C | N581S | missense_variant | 0.99 | 3 |
| 7:140781602:G | G469A | missense_variant | 1.00 | 2 |
| 7:140753355:T | D594N | missense_variant | 1.00 | 2 |
| 7:140781617:A | G464V | missense_variant | 1.00 | 1 |
| 7:140781611:G | G466A | missense_variant | 1.00 | 1 |
| 7:140781611:T | G466E | missense_variant | 1.00 | 1 |
| 7:140753324:A | W604L | missense_variant | 1.00 | 1 |
| 7:140753355:G | D594H | missense_variant | 1.00 | 1 |
| 7:140753349:T | G596S | missense_variant | 1.00 | 1 |
| 7:140753334:C | K601E | missense_variant | 1.00 | 1 |
| 7:140781603:A | G469* | stop_gained | 0.99 | 1 |
| 7:140753332:A | K601N | missense_variant | 0.99 | 1 |
| 7:140778054:G | L485S | missense_variant | 0.54 | 1 |
| 7:140781617:T | G464E | missense_variant | 1.00 | 0 |
| 7:140781602:T | G469E | missense_variant | 1.00 | 0 |
| 7:140781617:G | G464A | missense_variant | 1.00 | 0 |
| 7:140781603:G | G469R | missense_variant | 1.00 | 0 |
| 7:140781612:T | G466R | missense_variant | 1.00 | 0 |
| 7:140781612:G | G466R | missense_variant | 1.00 | 0 |
| 7:140781603:T | G469R | missense_variant | 1.00 | 0 |
| 7:140753324:G | W604S | missense_variant | 1.00 | 0 |
| 7:140753336:C | V600G | missense_variant | 1.00 | 0 |
| 7:140753325:C | W604G | missense_variant | 1.00 | 0 |
| 7:140753325:G | W604R | missense_variant | 1.00 | 0 |
| 7:140753325:T | W604R | missense_variant | 1.00 | 0 |
| 7:140753336:G | V600A | missense_variant | 1.00 | 0 |
| 7:140753349:G | G596R | missense_variant | 1.00 | 0 |
| 7:140753337:A | V600L | missense_variant | 1.00 | 0 |
| 7:140753337:G | V600L | missense_variant | 1.00 | 0 |
| 7:140753337:T | V600M | missense_variant | 1.00 | 0 |
| 7:140753348:A | G596V | missense_variant | 1.00 | 0 |
| 7:140753348:G | G596A | missense_variant | 1.00 | 0 |
| 7:140753348:T | G596D | missense_variant | 1.00 | 0 |
| 7:140753349:A | G596C | missense_variant | 1.00 | 0 |
| 7:140753355:A | D594Y | missense_variant | 1.00 | 0 |
| 7:140753333:G | K601T | missense_variant | 1.00 | 0 |
| 7:140753333:C | K601R | missense_variant | 1.00 | 0 |
| 7:140753333:A | K601I | missense_variant | 1.00 | 0 |
| 7:140753334:G | K601Q | missense_variant | 1.00 | 0 |
| 7:140753327:A | R603L | missense_variant | 1.00 | 0 |
| 7:140753327:G | R603P | missense_variant | 1.00 | 0 |
| 7:140753327:T | R603Q | missense_variant | 1.00 | 0 |
| 7:140781606:G | F468L | missense_variant | 1.00 | 0 |
| 7:140781606:T | F468I | missense_variant | 1.00 | 0 |
| 7:140781605:G | F468S | missense_variant | 1.00 | 0 |
| 7:140781605:C | F468C | missense_variant | 1.00 | 0 |
| 7:140781605:T | F468Y | missense_variant | 1.00 | 0 |
| 7:140781606:C | F468V | missense_variant | 1.00 | 0 |
| 7:140781609:C | S467A | missense_variant | 1.00 | 0 |
| 7:140781609:T | S467T | missense_variant | 1.00 | 0 |
| 7:140781609:G | S467P | missense_variant | 1.00 | 0 |
| 7:140781615:C | S465A | missense_variant | 1.00 | 0 |
| 7:140781615:T | S465T | missense_variant | 1.00 | 0 |
| 7:140781615:G | S465P | missense_variant | 1.00 | 0 |
| 7:140781612:A | G466* | stop_gained | 0.99 | 0 |
| 7:140753343:G | A598P | missense_variant | 0.99 | 0 |
| 7:140753343:T | A598T | missense_variant | 0.99 | 0 |
| 7:140753343:A | A598S | missense_variant | 0.99 | 0 |
| 7:140753354:C | D594G | missense_variant | 0.99 | 0 |
| 7:140753354:A | D594V | missense_variant | 0.99 | 0 |
| 7:140753354:G | D594A | missense_variant | 0.99 | 0 |
| 7:140753345:G | L597P | missense_variant | 0.99 | 0 |
| 7:140753345:C | L597R | missense_variant | 0.99 | 0 |
| 7:140753345:T | L597Q | missense_variant | 0.99 | 0 |
| 7:140753351:G | F595S | missense_variant | 0.99 | 0 |
| 7:140753352:C | F595V | missense_variant | 0.99 | 0 |
| 7:140753351:T | F595Y | missense_variant | 0.99 | 0 |
| 7:140753352:G | F595L | missense_variant | 0.99 | 0 |
| 7:140753352:T | F595I | missense_variant | 0.99 | 0 |
| 7:140753351:C | F595C | missense_variant | 0.99 | 0 |
| 7:140781608:A | S467L | missense_variant | 0.99 | 0 |
| 7:140753330:T | S602Y | missense_variant | 0.99 | 0 |
| 7:140753330:A | S602F | missense_variant | 0.99 | 0 |
| 7:140753330:C | S602C | missense_variant | 0.99 | 0 |
| 7:140781614:T | S465Y | missense_variant | 0.99 | 0 |
| 7:140781614:C | S465C | missense_variant | 0.99 | 0 |
| 7:140781614:A | S465F | missense_variant | 0.99 | 0 |
| 7:140753339:A | T599I | missense_variant | 0.99 | 0 |
| 7:140753339:T | T599K | missense_variant | 0.99 | 0 |
| 7:140753339:C | T599R | missense_variant | 0.99 | 0 |
| 7:140753342:C | A598G | missense_variant | 0.99 | 0 |
| 7:140753342:A | A598V | missense_variant | 0.99 | 0 |
| 7:140753342:T | A598D | missense_variant | 0.99 | 0 |
| 7:140753332:G | K601N | missense_variant | 0.99 | 0 |
| 7:140753340:A | T599S | missense_variant | 0.99 | 0 |
| 7:140753340:C | T599A | missense_variant | 0.99 | 0 |
| 7:140753340:G | T599P | missense_variant | 0.99 | 0 |
| 7:140753324:T | W604* | stop_gained | 0.99 | 0 |
| 7:140753353:C | D594E | missense_variant | 0.99 | 0 |
| 7:140753353:T | D594E | missense_variant | 0.99 | 0 |
| 7:140753393:A | N581I | missense_variant | 0.99 | 0 |
| 7:140753393:G | N581T | missense_variant | 0.99 | 0 |
| 7:140753334:A | K601* | stop_gained | 0.99 | 0 |
| 7:140753328:C | R603G | missense_variant | 0.99 | 0 |
| 7:140753331:G | S602P | missense_variant | 0.99 | 0 |
| 7:140753331:T | S602T | missense_variant | 0.99 | 0 |
| 7:140753331:C | S602A | missense_variant | 0.99 | 0 |
| 7:140753346:C | L597V | missense_variant | 0.99 | 0 |
| 7:140753346:T | L597I | missense_variant | 0.99 | 0 |
| 7:140781604:T | F468L | missense_variant | 0.99 | 0 |
| 7:140781604:C | F468L | missense_variant | 0.99 | 0 |
| 7:140753350:T | F595L | missense_variant | 0.98 | 0 |
| 7:140753350:C | F595L | missense_variant | 0.98 | 0 |
| 7:140781608:C | S467* | stop_gained | 0.98 | 0 |
| 7:140781608:T | S467* | stop_gained | 0.98 | 0 |
| 7:140753328:A | R603* | stop_gained | 0.97 | 0 |
| 7:140778054:C | L485W | missense_variant | 0.54 | 0 |
| 7:140778016:T | F498I | missense_variant | 0.51 | 0 |
| 7:140778016:G | F498L | missense_variant | 0.51 | 0 |
| 7:140778016:C | F498V | missense_variant | 0.51 | 0 |
| 7:140739819:C | F707C | missense_variant | 0.51 | 0 |
| 7:140778015:T | F498Y | missense_variant | 0.51 | 0 |
| 7:140739819:T | F707Y | missense_variant | 0.51 | 0 |
| 7:140778069:G | V480A | missense_variant | 0.51 | 0 |
| 7:140778069:C | V480G | missense_variant | 0.51 | 0 |
| 7:140778069:T | V480E | missense_variant | 0.51 | 0 |
| 7:140739819:G | F707S | missense_variant | 0.51 | 0 |
| 7:140778015:G | F498S | missense_variant | 0.51 | 0 |
| 7:140778063:T | V482E | missense_variant | 0.51 | 0 |
| 7:140778063:G | V482A | missense_variant | 0.51 | 0 |
| 7:140778063:C | V482G | missense_variant | 0.51 | 0 |
| 7:140778057:T | M484K | missense_variant | 0.51 | 0 |
| 7:140778057:G | M484T | missense_variant | 0.51 | 0 |
| 7:140778057:C | M484R | missense_variant | 0.51 | 0 |
| 7:140778015:C | F498C | missense_variant | 0.51 | 0 |
| 7:140778048:T | V487E | missense_variant | 0.51 | 0 |
| 7:140778048:G | V487A | missense_variant | 0.51 | 0 |
| 7:140778048:C | V487G | missense_variant | 0.51 | 0 |
| 7:140778024:G | L495S | missense_variant | 0.51 | 0 |
In silico saturation mutagenesis
| Cancer type | Selected cancer type | Mutational discovery Index | Feature Complexity |
|---|---|---|---|
| (LUAD) Lung adenocarcinoma | LUAD | 0.50 | 0.19 |