KRAS
ST
×
Select a mutation to see further information
| Mutation | AA change | Consequence type | BoostDM score | IntOGen frequency |
|---|---|---|---|---|
| 12:25245350:T | G12D | missense_variant | 1.00 | 9 |
| 12:25245350:A | G12V | missense_variant | 1.00 | 6 |
| 12:25245347:T | G13D | missense_variant | 1.00 | 5 |
| 12:25245351:A | G12C | missense_variant | 1.00 | 5 |
| 12:25245351:T | G12S | missense_variant | 1.00 | 4 |
| 12:25227341:G | Q61H | missense_variant | 0.94 | 2 |
| 12:25245350:G | G12A | missense_variant | 1.00 | 1 |
| 12:25245351:G | G12R | missense_variant | 1.00 | 1 |
| 12:25245348:G | G13R | missense_variant | 1.00 | 0 |
| 12:25245348:T | G13S | missense_variant | 1.00 | 0 |
| 12:25245347:G | G13A | missense_variant | 1.00 | 0 |
| 12:25245348:A | G13C | missense_variant | 1.00 | 0 |
| 12:25245347:A | G13V | missense_variant | 1.00 | 0 |
| 12:25245354:G | A11P | missense_variant | 0.99 | 0 |
| 12:25245354:A | A11S | missense_variant | 0.99 | 0 |
| 12:25245354:T | A11T | missense_variant | 0.99 | 0 |
| 12:25245345:A | V14L | missense_variant | 0.99 | 0 |
| 12:25245345:T | V14I | missense_variant | 0.99 | 0 |
| 12:25245345:G | V14L | missense_variant | 0.99 | 0 |
| 12:25227349:T | A59T | missense_variant | 0.99 | 0 |
| 12:25227340:G | E62Q | missense_variant | 0.99 | 0 |
| 12:25227340:T | E62K | missense_variant | 0.99 | 0 |
| 12:25227345:A | G60V | missense_variant | 0.99 | 0 |
| 12:25227345:G | G60A | missense_variant | 0.99 | 0 |
| 12:25227345:T | G60D | missense_variant | 0.99 | 0 |
| 12:25227346:A | G60C | missense_variant | 0.99 | 0 |
| 12:25227346:G | G60R | missense_variant | 0.99 | 0 |
| 12:25227346:T | G60S | missense_variant | 0.99 | 0 |
| 12:25227349:A | A59S | missense_variant | 0.99 | 0 |
| 12:25227349:G | A59P | missense_variant | 0.99 | 0 |
| 12:25245333:T | A18T | missense_variant | 0.98 | 0 |
| 12:25245333:A | A18S | missense_variant | 0.98 | 0 |
| 12:25245333:G | A18P | missense_variant | 0.98 | 0 |
| 12:25227338:G | E62D | missense_variant | 0.95 | 0 |
| 12:25227338:A | E62D | missense_variant | 0.95 | 0 |
| 12:25227341:A | Q61H | missense_variant | 0.94 | 0 |
| 12:25245337:A | K16N | missense_variant | 0.93 | 0 |
| 12:25245337:G | K16N | missense_variant | 0.93 | 0 |
| 12:25227340:A | E62* | stop_gained | 0.93 | 0 |
| 12:25245330:C | L19V | missense_variant | 0.93 | 0 |
| 12:25245330:T | L19M | missense_variant | 0.93 | 0 |
| 12:25245328:G | L19F | missense_variant | 0.93 | 0 |
| 12:25245328:A | L19F | missense_variant | 0.93 | 0 |
| 12:25227337:G | E63Q | missense_variant | 0.92 | 0 |
| 12:25227337:T | E63K | missense_variant | 0.92 | 0 |
| 12:25245341:T | G15D | missense_variant | 0.91 | 0 |
| 12:25245342:T | G15S | missense_variant | 0.91 | 0 |
| 12:25245342:G | G15R | missense_variant | 0.91 | 0 |
| 12:25245342:A | G15C | missense_variant | 0.91 | 0 |
| 12:25245341:G | G15A | missense_variant | 0.91 | 0 |
| 12:25245341:A | G15V | missense_variant | 0.91 | 0 |
| 12:25245335:T | S17N | missense_variant | 0.91 | 0 |
| 12:25245335:G | S17T | missense_variant | 0.91 | 0 |
| 12:25245335:A | S17I | missense_variant | 0.91 | 0 |
| 12:25227339:C | E62G | missense_variant | 0.91 | 0 |
| 12:25227339:A | E62V | missense_variant | 0.91 | 0 |
| 12:25227339:G | E62A | missense_variant | 0.91 | 0 |
| 12:25245353:A | A11V | missense_variant | 0.90 | 0 |
| 12:25245353:T | A11D | missense_variant | 0.90 | 0 |
| 12:25245353:C | A11G | missense_variant | 0.90 | 0 |
| 12:25227334:G | Y64H | missense_variant | 0.90 | 0 |
| 12:25227348:A | A59V | missense_variant | 0.90 | 0 |
| 12:25227348:C | A59G | missense_variant | 0.90 | 0 |
| 12:25227348:T | A59E | missense_variant | 0.90 | 0 |
| 12:25227334:C | Y64D | missense_variant | 0.90 | 0 |
| 12:25227334:T | Y64N | missense_variant | 0.90 | 0 |
| 12:25245338:A | K16M | missense_variant | 0.89 | 0 |
| 12:25245338:C | K16R | missense_variant | 0.89 | 0 |
| 12:25245339:G | K16Q | missense_variant | 0.89 | 0 |
| 12:25245338:G | K16T | missense_variant | 0.89 | 0 |
| 12:25245339:C | K16E | missense_variant | 0.89 | 0 |
| 12:25245319:A | Q22H | missense_variant | 0.89 | 0 |
| 12:25245319:G | Q22H | missense_variant | 0.89 | 0 |
| 12:25245329:G | L19S | missense_variant | 0.89 | 0 |
| 12:25245329:C | L19W | missense_variant | 0.89 | 0 |
| 12:25245332:A | A18V | missense_variant | 0.89 | 0 |
| 12:25245332:C | A18G | missense_variant | 0.89 | 0 |
| 12:25245332:T | A18D | missense_variant | 0.89 | 0 |
| 12:25245344:C | V14G | missense_variant | 0.89 | 0 |
| 12:25245344:G | V14A | missense_variant | 0.89 | 0 |
| 12:25245344:T | V14E | missense_variant | 0.89 | 0 |
| 12:25227335:A | E63D | missense_variant | 0.85 | 0 |
| 12:25227335:G | E63D | missense_variant | 0.85 | 0 |
| 12:25227342:C | Q61R | missense_variant | 0.84 | 0 |
| 12:25227342:G | Q61P | missense_variant | 0.84 | 0 |
| 12:25227342:A | Q61L | missense_variant | 0.84 | 0 |
| 12:25227343:T | Q61K | missense_variant | 0.83 | 0 |
| 12:25227343:C | Q61E | missense_variant | 0.83 | 0 |
| 12:25245322:C | I21M | missense_variant | 0.81 | 0 |
| 12:25245334:C | S17R | missense_variant | 0.81 | 0 |
| 12:25245334:T | S17R | missense_variant | 0.81 | 0 |
| 12:25227337:A | E63* | stop_gained | 0.71 | 0 |
| 12:25245339:A | K16* | stop_gained | 0.67 | 0 |
| 12:25245329:T | L19* | stop_gained | 0.65 | 0 |
| 12:25227336:A | E63V | missense_variant | 0.62 | 0 |
| 12:25227336:G | E63A | missense_variant | 0.62 | 0 |
| 12:25227336:C | E63G | missense_variant | 0.62 | 0 |
| 12:25245327:G | T20P | missense_variant | 0.59 | 0 |
| 12:25245327:A | T20S | missense_variant | 0.59 | 0 |
| 12:25245336:A | S17C | missense_variant | 0.59 | 0 |
| 12:25245336:C | S17G | missense_variant | 0.59 | 0 |
| 12:25245336:G | S17R | missense_variant | 0.59 | 0 |
| 12:25245324:G | I21L | missense_variant | 0.59 | 0 |
| 12:25245324:C | I21V | missense_variant | 0.59 | 0 |
| 12:25245324:A | I21L | missense_variant | 0.59 | 0 |
| 12:25245320:G | Q22P | missense_variant | 0.59 | 0 |
| 12:25245320:C | Q22R | missense_variant | 0.59 | 0 |
| 12:25245320:A | Q22L | missense_variant | 0.59 | 0 |
| 12:25245315:G | I24L | missense_variant | 0.59 | 0 |
| 12:25245315:C | I24V | missense_variant | 0.59 | 0 |
| 12:25245315:A | I24F | missense_variant | 0.59 | 0 |
| 12:25245327:C | T20A | missense_variant | 0.59 | 0 |
| 12:25245318:T | L23I | missense_variant | 0.59 | 0 |
| 12:25245318:C | L23V | missense_variant | 0.59 | 0 |
| 12:25245317:G | L23P | missense_variant | 0.57 | 0 |
| 12:25245317:C | L23R | missense_variant | 0.57 | 0 |
| 12:25245323:C | I21R | missense_variant | 0.57 | 0 |
| 12:25245323:G | I21T | missense_variant | 0.57 | 0 |
| 12:25245323:T | I21K | missense_variant | 0.57 | 0 |
| 12:25245326:A | T20M | missense_variant | 0.57 | 0 |
| 12:25245326:C | T20R | missense_variant | 0.57 | 0 |
| 12:25245326:T | T20K | missense_variant | 0.57 | 0 |
| 12:25245314:G | I24T | missense_variant | 0.57 | 0 |
| 12:25245321:T | Q22K | missense_variant | 0.57 | 0 |
| 12:25245317:T | L23Q | missense_variant | 0.57 | 0 |
| 12:25245314:C | I24S | missense_variant | 0.57 | 0 |
| 12:25245314:T | I24N | missense_variant | 0.57 | 0 |
| 12:25245321:C | Q22E | missense_variant | 0.57 | 0 |
| 12:25227343:A | Q61* | stop_gained | 0.56 | 0 |
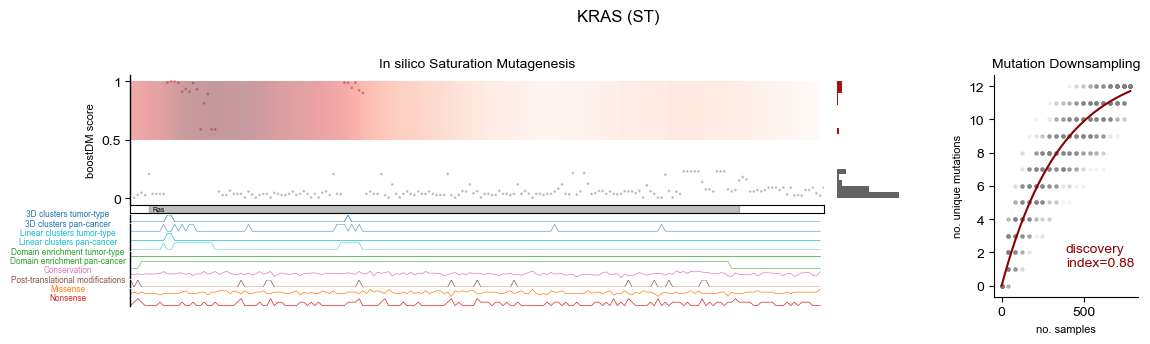
In silico saturation mutagenesis
| Cancer type | Selected cancer type | Mutational discovery Index | Feature Complexity |
|---|---|---|---|
| (ST) Stomach adenocarcinoma | ST | 0.88 | 0.14 |